There is considerable excitement about graphene-based biosensors. In particular, the material’s unique structure and electronic properties offers great potential for rapid, reliable DNA/RNA sensing and sequencing.
To date, this potential has been checked by a lack of fundamental understanding of graphene−nucleobase interactions and the origin of measured molecular fingerprints.
A recent study has defined key interactions as DNA/RNA nucleobases are adsorbed onto graphene, and identified specific improvements to current graphene-based biosensing, to improve sensitivity.
The research, led by Monash University’s Yuefeng Yin and FLEET chief investigator Nikhil Medhekar, with Jiri Cervenka at the Czech Institute of Physics (Academy of Sciences of the Czech Republic), was a systematic study of the role of the surface coverage and adsorption geometry on the electronic structure of DNA/RNA nucleobases on graphene.
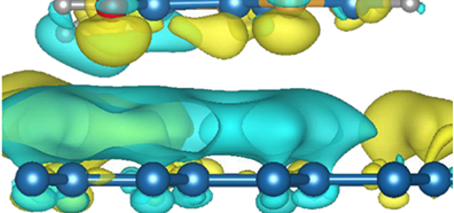
Studying the electronic structure of nucleic acid bases on graphene as a function of molecular orientation and surface coverage, the researchers found that the adsorption behavior of the nucleobases on graphene is determined by a subtle balance between graphene−molecule and intermolecular interactions, where the resulting adsorption geometry and energetics of the system are often decided by the interaction with surrounding molecules, leading to a tilt of the nucleobases on graphene at high molecular densities.
As the nucleobases possess strong in-plane dipole moments, their tilt causes significant changes of the electronic structure of the whole graphene−nucleobase system.
The study explored the possibility of enhancing electronic fingerprints of nucleobases adsorbed on graphene by tuning the surface coverage and modifying (maximising) molecular dipoles.
The discovery that in-plane dipole moments play a dominant role in altering the electronic structure of graphene suggests new strategies for improved DNA/RNA detection, using vertically aligned nucleobases adsorbed on graphene.
These results highlight and provide critical fundamental knowledge for the identification of molecular fingerprints in graphene-based nucleobase sensing and sequencing methods.
The results provide new understanding of interfacial interactions and molecule-specific signatures of DNA/RNA nucleobases on graphene, as well as specific opportunities to improve the design of current electrochemical sensing devices. This has exciting potential for graphene-based biosensors and end-users in fields from medicine to forensics to technology.
The work was supported by the ARC Discovery Project as well as by FLEET, the Monash Campus Cluster, National Computer Institute and the Pawsey Supercomputing facility. The study was published in the Journal of Physical Chemistry Letters in June 2017.
Want to learn more? You can follow FLEET on Twitter, watch a video on future solutions to computation energy use, or subscribe to FLEET news.